Genetic Basis of Cancer
DNA
[See 1.2 DNA & the Central Dogma]
DNA stands for deoxyribonucleic acid. DNA is made of two strands, called polynucleotides, that coil around each other to form a double helix. Each polynucleotide strand is a long chain composed of many individual units, called nucleotides.
Each nucleotide has three parts: a monosaccharide called deoxyribose, a phosphate group, and a nitrogenous base. There are four types of nucleotides: guanine (G), cytosine (C), adenine (A) and thymine (T) – these four each have a different nitrogenous base component.
The arrangement of these four bases along the strand is what is known as the DNA sequence, and is the basis for life, spelling out genes which encode for every protein in our body.
In the DNA strand, the nucleotides are joined to one another by phosphodiester bonds. The two strands are then joined to one another to form a double-stranded DNA molecule by hydrogen bonds between the bases of each strand. G always hydrogen bonds with C, and A always hydrogen bonds with T.
The human genome (which consists of approximately 3.2 billion nucleotides) is distributed over 23 pairs of chromosomes, which include 22 pairs of autosomes and 2 sex chromosomes. Each chromosome is one very long double-stranded DNA molecule, which is coiled incredibly tightly and wound around special proteins called histones. Because DNA is negatively charged, it is able to closely bind to these positively-charged histones. DNA can coil so tightly that a strand 2 metres long can be packaged into one cell nucleus, which has a diameter of just 6 um!
The complex of DNA and histones is called chromatin. The basic repeating unit of chromatin is called the nucleosome, which consists of a short segment of DNA wrapped around 8 histones (a histone octamer).
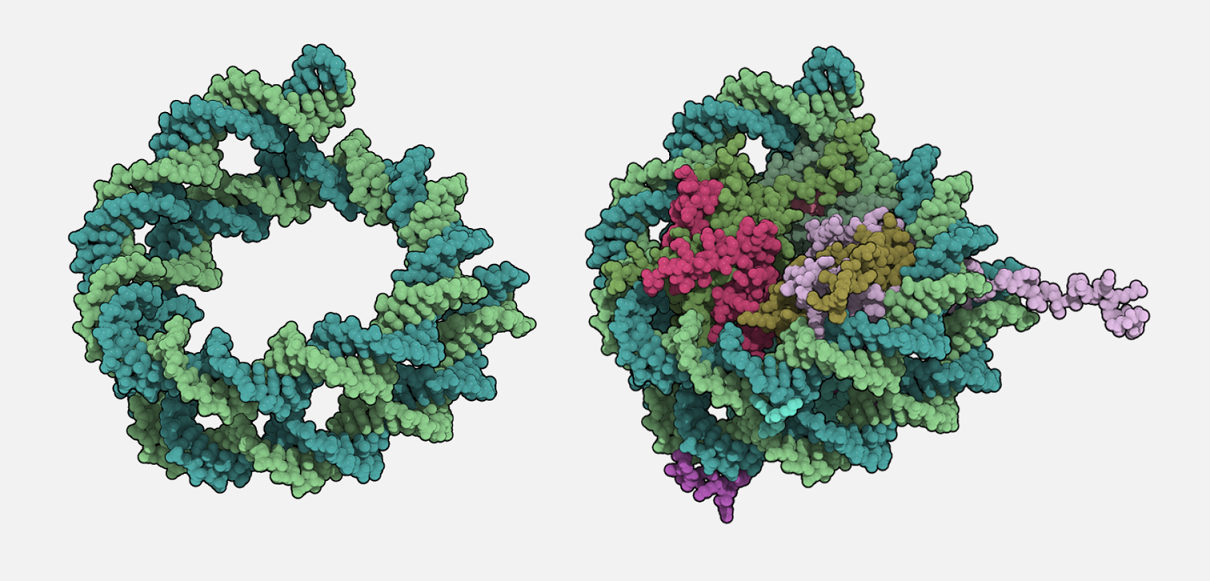
RIGHT That same coil of DNA, but wrapped around the positively-charged histones to form a nucleosome.
Mutations
A genetic mutation is a change in the nucleotide sequence of the genome. The effect of a genetic mutation depends on where it occurs: mutations affecting the function of an essential protein are more likely to cause severe disease or cancer.
A point mutation is a change in a single nucleotide. Point mutations are often caused by mutagens. A point mutation may or may not change the amino acid sequence of a protein. This is generally attributed to the "wobble effect" of the codons that are translated into amino acids. Point mutations are often referred to as single nucleotide polymorphisms.
- A silent mutation is a point mutation that results in the production of a protein that is either identical or very similar to the original protein. Generally, these mutations are non-deleterious, i.e. not harmful. In some cases, these mutations may even be advantageous.
See this paper for a recent example of advantageous silent mutations.
A missense mutation is a point mutation that results in the production of a protein that is different to the original protein. Generally, these are deleterious and result in functional loss of the protein. For example, in Sickle Cell Anemia, a point mutation converts a GAG codon into a GUG codon. Translation of the mutated protein now encodes for the amino acid valine instead of glutamic acid. Ultimately, there is a functional deficit in hemoglobin.
A nonsense mutation is a point mutation that results in the production of a truncated protein: it causes translation to stop partway along the protein’s length, so that the protein cannot be completed, resulting in a near-total (or total) loss of function.
Duchenne Muscular Dystrophy (DMD) is a genetic disease with various genetic causes, e.g. deletions, nonsense mutations. In nonsense mutation variants, the truncated protein may result in the loss of function of the protein N-terminus, which is important for binding between the muscle fiber sarcolemma and the actin thin filament. DMD patients suffer from loss of mobility, due to the weakened muscle architecture, impaired contraction, inflammation, and fibrosis.
A deletion is the removal of one or more nucleotides.
An insertion is the addition of one or more extra nucleotides. Insertions are often caused by transposable elements, which are stretches of DNA that are able to “jump” around different points in the genome.
Deletions and insertions are particularly dangerous, as these are extremely likely to change the reading frame of mRNA sequence. Consider the DNA sequence:
[AAT][CCC][GCC][TTT][ACC][ACC][ATA][G..]
and the resulting amino acid sequence:
Leu-Gly-Arg-Lys-Trp-Trp-Tyr-...
A deletion of the bolded T will result the DNA sequence:
[AAT][CCC][GCC][TTA][CCA][CCA][TAG]
which translates to:
Leu-Gly-Arg-Asn-Gly-Gly-Ile...
Note there is a complete transformation in amino acid sequence after Arg. This is extremely likely to lead to a non-functional protein.
A duplication is a increase in the number of a discrete nucleotide sequence (e.g. AAGTT --> AAGTTAAGTT - note the 2 AAGTT sequences). Generally, a duplication manifests as an increase in the copy number of exons (protein-coding segments of the human genome).
While deceptively similar to an insertion, there is a difference:
- An insertion is just an "addition" of one or more nucleotides. A discrete nucleotide sequence/exon is NOT copied, whether the insertion is due to error-prone DNA replication or the movement of transposable elements.
- A duplication results in an increase in the copy number of some discrete nucleotide sequence (most likely an exon). The exact cause (or causes) of an event are not fully understood.
This paper from 2001 explores a relationship between alternative splicing and exon duplication, and how this may generate functionally diverse proteins.
Because a duplication does ultimately alter the nucleotide sequence, there is potential for disrupted gene function. Listed below are examples of conditions that have been associated with duplications.
Click the condition to read the associated research paper/abstract.
What has been discussed are changes on a nucleotide or segment level. However there can be genetic changes on the level of chromosomes. Neochromosomes are giant chromosomes resulting from the fusion of DNA from many different fragments of others chromosomes. These neochromosomes may contain elevated numbers of oncogenes e.g. MDM2. Such structures are rare overall (~3% of cancers) but can be common in certain cancers such as liposarcomas.
Which of the following statements about mutations is correct?
DNA Damage & Repair
DNA is everything to a cell, without a correct version of it, a cell cannot function. After all, it is from DNA that mRNA is transcribed and then translated into proteins – the functional machinery of cells. It can be analogous to our dependence on water. We need water to survive. Without it, we’d die of dehydration. But we can’t just drink any type of water – it needs to be clean. Contaminated water, would just end up killing us – either slowly, or depending on toxins, extremely quickly. This same idea is evident in cells – mutations ‘contaminate’ the integrity of DNA. Some mutations are more deleterious or harmful than others. The time in which mutations are most likely to occur is during S phase, when DNA is replicating. Hence, the effect of carcinogens is most strongly felt here. To protect itself from mutations and DNA damage, the cell has developed mechanisms that enable it to recognise and repair DNA.
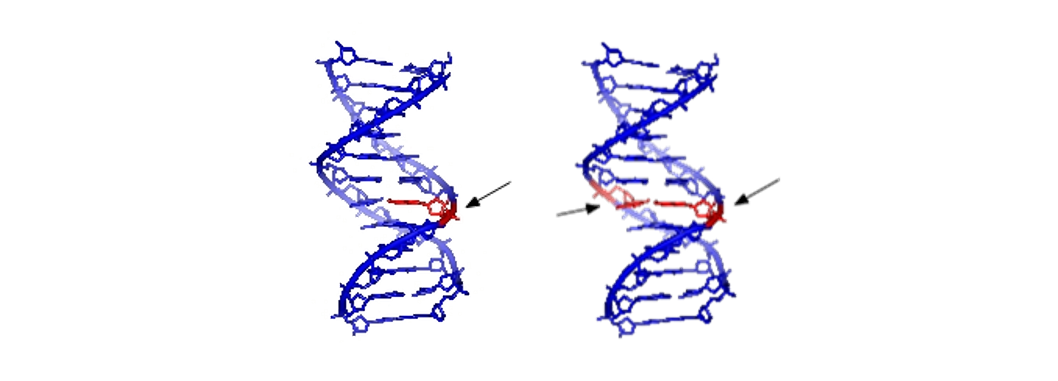
RIGHT Double stand DNA damage – both sides will require excision.
—
Single stranded and double stranded damage to DNA / CC-BY-SA-3.0
There are four main ways that DNA can be damaged:
A base is missing.
A nucleotide on one strand is damaged.
Incorrect pairing has happened.
Both strands are damaged.
The repair of DNA in these four cases, listed from least to greatest in complexity, involves various enzymes that: recognise the damage, cut out a portion of the DNA if needed and replace the damaged DNA with correct bases. However, in some cases the damage is too great to fix. In these situations, a back-up mechanism kicks in. The accumulation of DNA damage is detected by other proteins that then trigger apoptosis – cell death. This shows how important DNA integrity is – the cell will undergo apoptosis in the cases where DNA damage can’t be fixed. It is essentially a sacrifice for the greater good – evolution has recognised the importance of removing damaged cells to prevent progression to states that are harmful to the organism, like cancer.
In all cancers, the recognition of DNA damage, or the process of apoptosis has been affected. Cancer cells have developed their own mechanisms that override the back-up systems of cells, ultimately leading to their proliferation.
Tumour Suppressor Genes
A tumour suppressor gene encodes a protein that inhibits cell proliferation. They are an important part of a cell’s healthy function. You can think of tumour suppressors like the brakes on a car, which can slow the cell down or bring it to a stop to avoid danger. If these genes stop functioning, just like a car with no brakes, the cell will begin to lose control – and turn into a tumour.
Because genes exist in two copies (two alleles), both copies of a tumour suppressor gene must be lost for its function to be completely stopped. If the front brakes of your car are broken, the car is still able to stop using the rear brakes – though it won’t be quite as effective. That is, two loss-of-function mutations are required in a tumour suppressor gene to turn a cell cancerous. There are, however, certain tumour suppressor gene mutations that only require ONE loss-of-function mutation. These are called dominant-negative mutations.
The first mutation may be inherited (germinal), or develop spontaneously within a person’s lifetime (somatic). The second mutation is almost always somatic. If someone is born with one inherited mutation, they will not have cancer straight away – but they are much more likely to develop cancer during their lifetime, as they only need one more mutation.
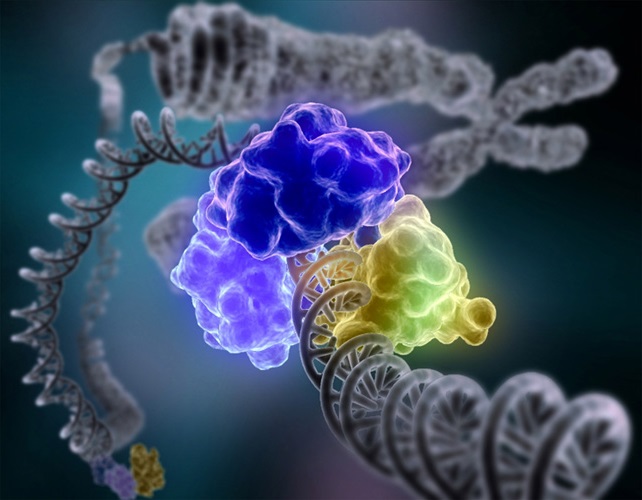
RIGHT A p53 model bound to a strand of DNA.
—
David S. Goodsell & the RCSB PDB / CC BY 4.0
The most important example of a tumour suppressor gene is p53, which comes into play when the cell detects DNA damage. When damage occurs, p53 is activated and stops cell division until the DNA has been repaired [see DNA repair section]. If the damage can’t be repaired, p53 will induce programmed cell death (apoptosis) – the cell will die, preventing it from potentially growing into a tumour. The loss of p53 means that cells with damaged DNA can continue to divide, which can eventually lead to cancer. p53 is the most commonly mutated gene in human cancer cases. In fact, it’s been called the “guardian of the genome” because of how vitally important its protective role is!
Did you know that big animals like elephants and whales only rarely get cancer? This is pretty surprising, because an elephant has many more cells than a human due to its huge size – this should mean that cancer-causing mutations are far more likely to happen! However, researchers recently discovered that elephants actually have 20 copies of p53 in their genome, while humans only have one. That’s how elephants are able to avoid cancer so well!
Oncogenes
A proto-oncogene is a gene that encodes a protein that promotes cell proliferation. These genes are normal, and part of a healthy cell’s function. However, sometimes they can be mutated and become over-active. If this happens, it becomes known as an oncogene. Only a mutation in one copy of the proto-oncogene is needed to convert it into an oncogene. This is unlike tumour suppressor genes, which require two mutations to become cancerous (exception: see dominant-negative mutations in Tumor Suppressor Genes section).
![Illustration of how a normal cell is converted to a cancer cell, when an oncogene becomes activated.<br />—<br />Oncogenes from National Cancer Institute [Public Domain] / <a href="https://creativecommons.org/publicdomain/mark/1.0/">CC-PD-Mark</a>](https://upload.wikimedia.org/wikipedia/commons/thumb/d/de/Oncogenes_illustration.jpg/1280px-Oncogenes_illustration.jpg)
—
Oncogenes from National Cancer Institute [Public Domain] / CC-PD-Mark
The two most important examples of proto-oncogenes are the genes encoding for Ras and Myc. In both cases mutations can lead to constitutive signalling of the Ras and Myc proteins, causing the cell to move through G1, pass the G1 checkpoint, and enter the S phase, in an unregulated manner. In the case of Ras, mutations in the Ras gene can lead to changes in the structure of the Ras protein so that it is resistant to signals that turn it off. This means that Ras will be always activated, and hence its signalling is continuous. On the other hand, mutations in the Myc gene can lead to excessive production of the Myc protein. This will constantly stimulate the cell to pass the G1 checkpoint.
Which of the following statements about tumor suppressor genes and oncogenes is correct?
Epigenetics
The mutations we’ve already looked at are all changes in the actual sequence of the DNA. DNA sequence is a bit like a language, with genes being the words or sentences that tell the cell what to do. A genetic mutation causes the “words” to become scrambled and turn into nonsense.
However, the words themselves aren’t the whole story: for example, you might underline or highlight specific words in a sentence to remind yourself that they’re important. This is similar to what a cell does when it makes epigenetic changes: it can add special chemical groups around the DNA, which are like side-notes that signal if that gene should be transcribed more, or even turned off completely. But the actual words, or DNA sequences, don’t change!
Epigenetics refers to changes in gene function that are not caused by changes in DNA sequence. Importantly, they are still mitotically heritable: an epigenetic change is passed on to the cell’s daughters when it divides.
The word epigenetics comes from Greek, meaning “on top of” the gene.
There are a lot of different epigenetic changes that can occur, and there’s still a lot for researchers to discover about them. Some of them can have profound effects relating to cancer, as it’s been found that cancer cells have a very different epigenetic profile to healthy cells.
One type of modification found in cancer cells is the DNA methylation of certain parts of DNA, often at gene promoters. Methylation at a promoter region causes silencing of that gene – so it won’t get transcribed or translated into protein. In cancer cells, this type of epigenetic silencing happens at tumour suppressor genes. This releases the brakes and contributes to its ability to grow out of control and be cancerous.
Nature provides some more information on epigenetic influences.
![Epigenetic Mechanisms by National Institutes of Health [Public Domain] / <a href="https://creativecommons.org/publicdomain/mark/1.0/">CC-PD-Mark</a>](https://upload.wikimedia.org/wikipedia/commons/d/dd/Epigenetic_mechanisms.jpg)
This paper discusses an example of epigenetic mutations, specifically histone mutations, and their significance in DIPG (diffuse intrinsic pontine gliomas), a type of brain cancer with no current effective therapy.
Which of the following statements about epigenetics is correct?
Models of Cancer Progression
Researchers have developed two models that describe how cancer forms, the first being the linear progression model.
This model essentially describes how a cancer can form from the accumulation of mutations in one cell. For example:
- An epithelial cell in the small intestine has a mutation in a tumour suppressor gene. This mutation may have occurred randomly, or the cell may have inherited the mutation (a germline mutation).
- The cell gains another mutation in the same tumour suppressor gene, perhaps from radiation. This means that the gene is now non-functional and the cell has lost part of its regulatory mechanism.
- The cell then starts proliferating into a benign tumour. Because it can proliferate at a faster rate, the cell takes over its area, absorbing more nutrients and causing damage to nearby healthy cells.
- Some time later, another event occurs and this time the cell gains a mutation in a proto-oncogene. This means that not only has the cell lost a suppressive regulatory mechanism, but now, proliferation is promoted.
- If the cell then obtains mutations in both copies of p53, it becomes a carcinoma and in this case, colon cancer develops.
So, as you can see, the linear progression model describes how a cell slowly becomes this monstrous entity that, through mutations, gradually obtains more and more characteristics, which enables it to overwhelm and invade surrounding tissues.
But why is it that someone can have multiple cancers at a given time – surely the chances of having so many mutations randomly appearing is extremely low? The parallel progression model answers this question.
This model involves stem cells. Stem cells are special cells that can divide and become different cell types. For example, stem cells in the bone marrow can differentiate into different red blood cells AND white blood cells. Embryonic stem cells are cells that can differentiate to form any tissue in the body.
Now, imagine you have a stem cell and it has an inherited mutation in one of its tumour suppressor genes. This cancerous stem cell then proliferates and differentiates into many different cell types that can make up: breast tissue, liver tissue, lung tissue and bone. This means that in all of these tissues they are cells that have already have a mutation in one of its tumour suppressor gene. So only one more mutation is required for these cells to gain cancerous properties. In this model, there is generally one primary tumour and several secondary tumours. However, because the cells differ in cell types, all of these tumours would have different characteristics, making this situation extremely difficult to treat.

A major focus of cancer immunotherapy aims to address the secondary tumors that arise in the parallel progression model. Since these secondary tumors characterize metastatic disease, treatment of the primary tumour is nowhere near sufficient enough to improve disease prognosis.
For your interest, here's a recent paper about immunotherapy research, from the reputable Nature journal, authored by researchers at Harvard Medical School in the US.
Which of the following statements about models of cancer progression is correct?